---
title : 'The EM algorithm for Mixture Models'
subtitle : 'Bayesian Statistics: Mixture Models'
categories:
- Bayesian Statistics
keywords:
- Mixture Models
- Homework Honors
---
## Infer parameter of mixture of exponential and long normal for lifetime of fuses
::: {.callout-note}
### Instructions
Data on the lifetime (in years) of fuses produced by the ACME Corporation is available in the file fuses.csv:
In order to characterize the distribution of the lifetimes, it seems reasonable to fit to the data a two-component mixture of the form:
$$
f(x)=wλexp{−λx}+(1−w) \frac{1}{\sqrt{2\pi}\tau x} \exp{− \frac{(log(x)−μ)^{2}}{2τ^{2}}}, \quad x > 0.
$$ {#eq-mixture}
where $w$ is the weight associated with the exponential distribution, $\lambda$ is the rate of the exponential distribution, and $\text{LN}(\mu, \tau)$ is a log-normal distribution with mean $\mu$ and standard deviation $\tau$.
- Modify code to Generate n observations from a mixture of two Gaussian # distributions into code to sample 100 random numbers from a mixture of 4 exponential distributions with means 1, 4, 7 and 10 and weights 0.3, 0.25, 0.25 and 0.2, respectively.
- Use these sample to approximate the mean and variance of the mixture.
:::
```{r}
#| label: lbl-load-data
## Clear the environment and load required libraries
rm(list=ls())
set.seed(81196) # So that results are reproducible (same simulated data every time)
# Load the data
fuses <- read.csv("data/fuses.csv",header=FALSE)
x <- fuses$V1
n <- length(x) # Number of observations
# how many rows in the data
nrow(fuses)
# how many zeros in x
sum(x==0)
# almost half of the data is zeros!
par(mfrow=c(1,1))
xx.true = seq(0, max(x), by=1)
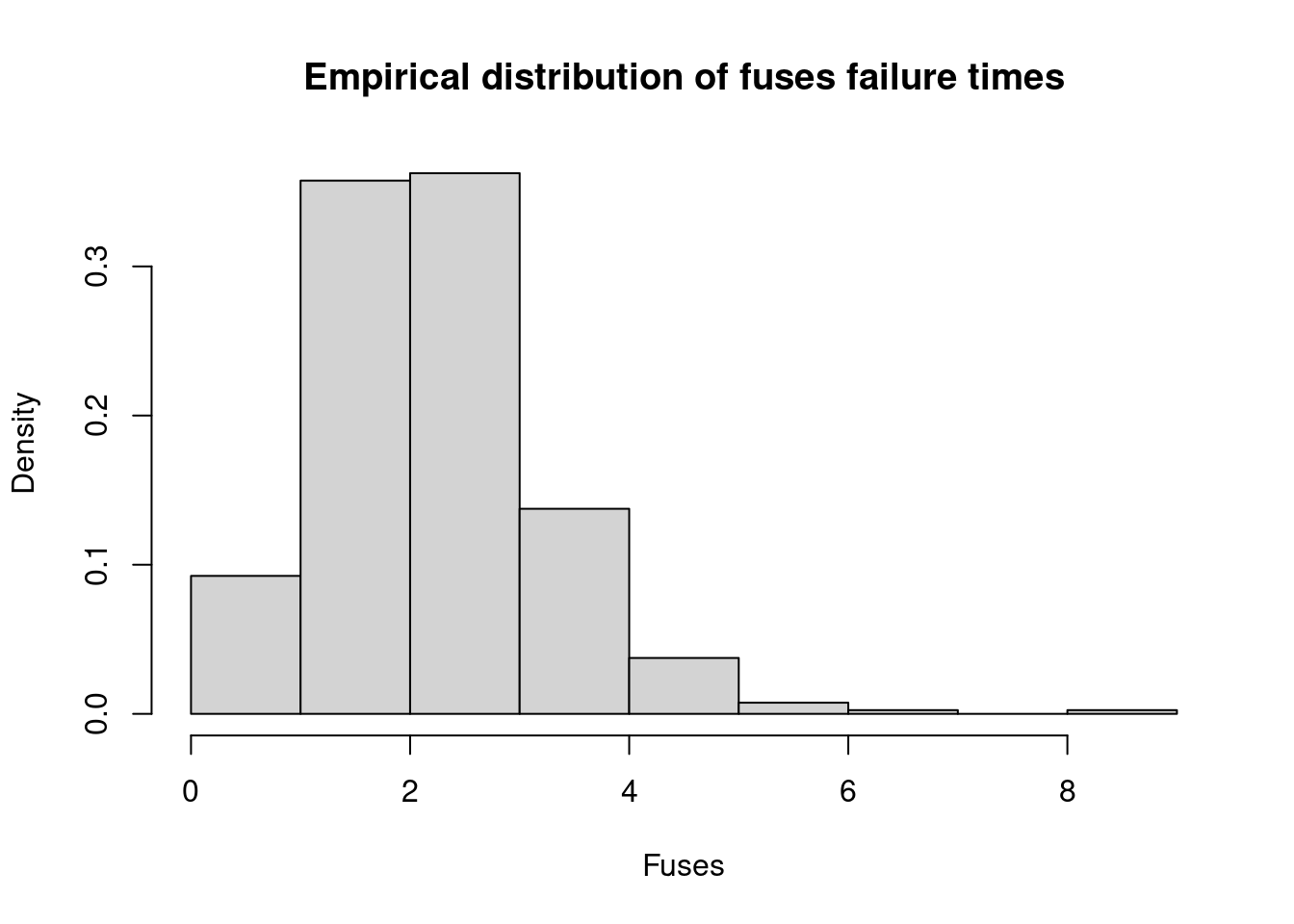
hist(x, freq=FALSE, xlab="Fuses", ylab="Density", main="Empirical distribution of fuses failure times")
```
```{r}
#| label: em_algorithm_loop
KK = 2 # Number of components
w = 0.05 # Assign equal weight to each component to start with
#mu = rnorm(1,mean(log(x)), sd(log(x)))
mu = mean(log(x))
tau = sd(log(x))
lambda = 20 / mean(x)
s = 0 # s_tep counter
sw = FALSE # sw_itch to stop the algorithm
QQ = -Inf # the Q function (log-likelihood function)
QQ.out = NULL # the Q function values
epsilon = 10^(-5) # the stopping criterion for the algorithm
trace <- data.frame(iter=0, w=w, lambda=lambda, mu=mu, tau=tau)
while(!sw){ ##Checking convergence
## E step
v = array(0, dim=c(n,KK))
for(i in 1:n){
v[i,1] = log(w) + dexp(x[i], rate=lambda, log=TRUE)
v[i,2] = log(1-w) + dlnorm(x[i], mu, tau, log=TRUE)
v[i,] = exp(v[i,] - max(v[i,]))/sum(exp(v[i,] - max(v[i,])))
}
## M step
w = mean(v[,1]) # Weights
lambda = sum(v[,1]) / sum(v[,1] * x) # Lambda (rate)
mu = sum(v[,2] * log(x)) / sum(v[,2]) # Mean
tau = sqrt(sum(v[,2] * (log(x) - mu)^2) / sum(v[,2])) # Tau (standard deviation)
# collect trace of parameters
trace = rbind(trace, data.frame(iter=s, w=w, lambda=lambda, mu=mu, tau=tau))
## Check convergence
QQn = 0
#vectorized version
log_lik_mat = v[,1]*(log(w) + dexp(x, lambda, log=TRUE)) +
v[,2]*(log(1-w) + dlnorm(x, mu, tau, log=TRUE))
QQn = sum(log_lik_mat)
if(abs(QQn-QQ)/abs(QQn)<epsilon){
sw=TRUE
}
QQ = QQn
QQ.out = c(QQ.out, QQ)
s = s + 1
print(paste(s, QQn))
}
```
next report the MLE parameters of the model.
```{r}
#| label: lbl-mle-estimates
# Report the MLE parameters
cat("w =", round(w, 2), "lambda =", round(lambda, 2), "mu =", round(mu, 2),"tau =", round(tau, 2))
```