polar2screen <- function( dist, origin, theta ) {
## takes dist, angle and origin and returns x and y of destination point
vx <- cos(theta) * dist;
vy <- sin(theta) * dist;
c( origin[1]+vx , origin[2]+vy );
}
screen2polar <- function( origin, dest ) {
## takes two points and returns distance and angle, from origin to dest
vx <- dest[1] - origin[1];
vy <- dest[2] - origin[2];
dist <- sqrt( vx*vx + vy*vy );
theta <- asin( abs(vy) / dist );
## correct for quadrant
if( vx < 0 && vy < 0 ) theta <- pi + theta; # lower-left
if( vx < 0 && vy > 0 ) theta <- pi - theta; # upper-left
if( vx > 0 && vy < 0 ) theta <- 2*pi - theta; # lower-right
if( vx < 0 && vy==0 ) theta <- pi;
if( vx==0 && vy < 0 ) theta <- 3*pi/2;
## return angle and dist
c( theta, dist );
}
point.polar <- function(dist,theta,origin=c(0,0),...) {
# angle theta is in radians
pt <- polar2screen(dist,origin,theta)
points( pt[1] , pt[2] , ... )
invisible( pt )
}
line.polar <- function(dist,theta,origin=c(0,0),...) {
# dist should be vector of length 2 with start and end points
# theta is angle
pt1 <- polar2screen(dist[1],origin,theta)
pt2 <- polar2screen(dist[2],origin,theta)
lines( c(pt1[1],pt2[1]), c(pt1[2],pt2[2]) , ... )
}
line.short <- function(x,y,short=0.1,...) {
# shortens the line segment, but retains angle and placement
pt1 <- c(x[1],y[1])
pt2 <- c(x[2],y[2])
theta <- screen2polar( pt1 , pt2 )[1]
dist <- screen2polar( pt1 , pt2 )[2]
q1 <- polar2screen( short , pt1 , theta )
q2 <- polar2screen( dist-short , pt1 , theta )
lines( c(q1[1],q2[1]) , c(q1[2],q2[2]) , ... )
}
wedge <- function(dist,start,end,pt,hedge=0.1,alpha,lwd=2,...) {
# start: start angle of wedge
# end: end angle of wedge
# pt: vector of point bg colors
n <- length(pt)
points.save <- matrix(NA,nrow=n,ncol=2) # x,y columns
span <- abs(end-start)
span2 <- span*(1 - 2*hedge)
origin <- start + span*hedge
gap <- span2/n
theta <- origin + gap/2
border <- rep("black",length(pt))
if ( !missing(alpha) ) {
pt <- sapply( 1:length(pt) , function(i) col.alpha(pt[i],alpha[i]) )
border <- sapply( 1:length(pt) , function(i) col.alpha(border[i],alpha[i]) )
}
for ( i in 1:n ) {
points.save[i,] <- point.polar( dist , theta , pch=21 , lwd=lwd , bg=pt[i] , col=border[i] , ... )
theta <- theta + gap
}
points.save
}
#######
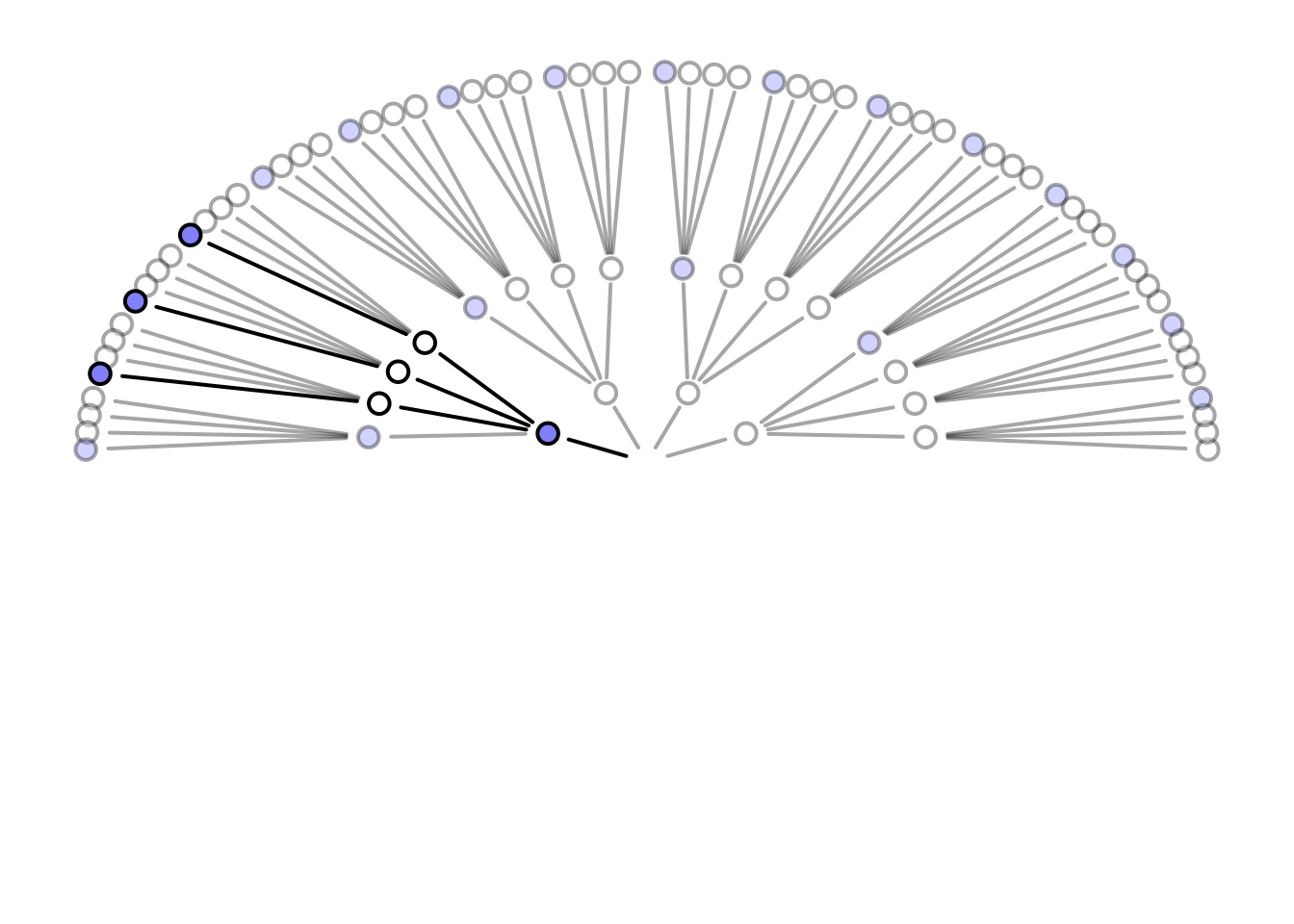
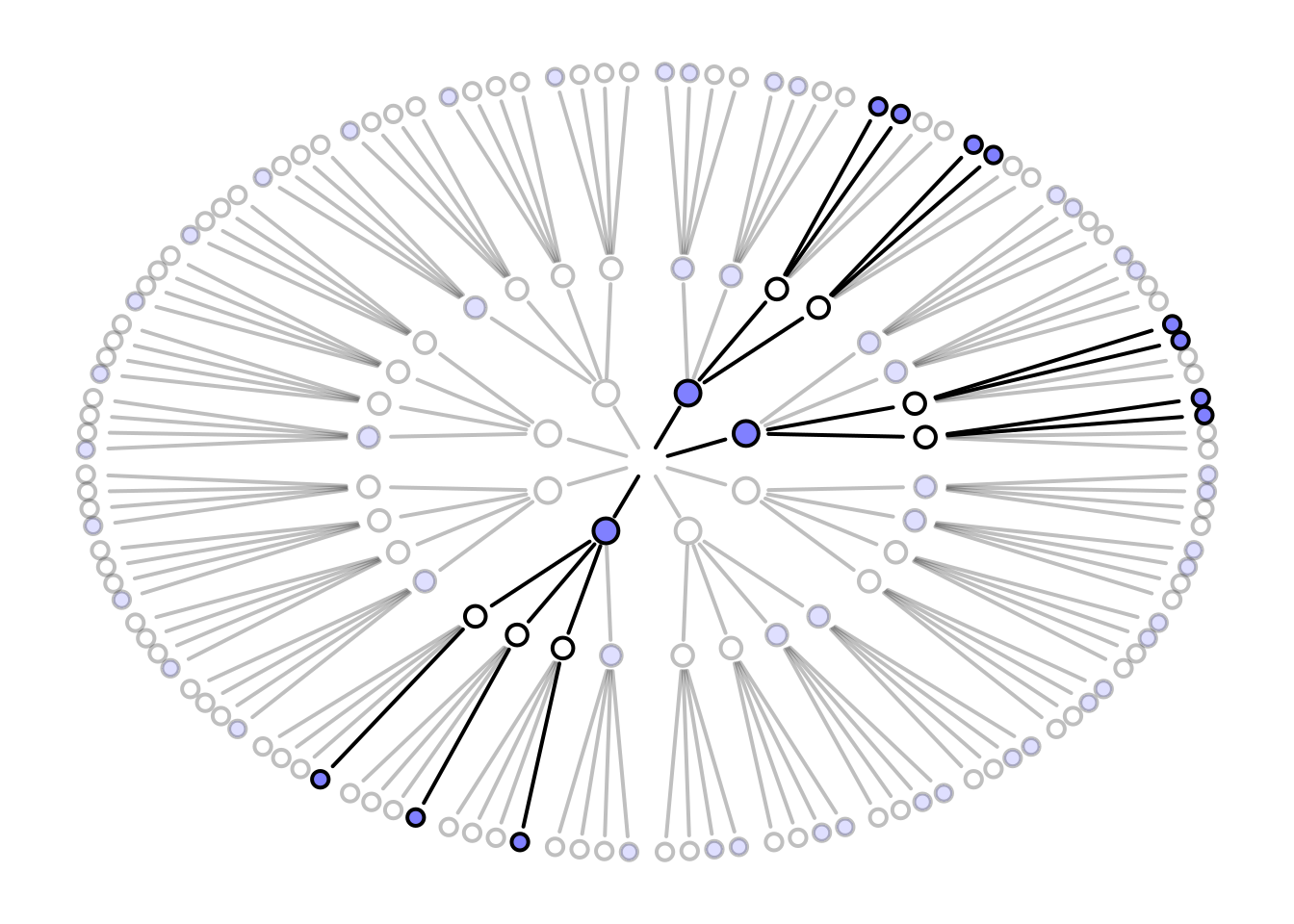
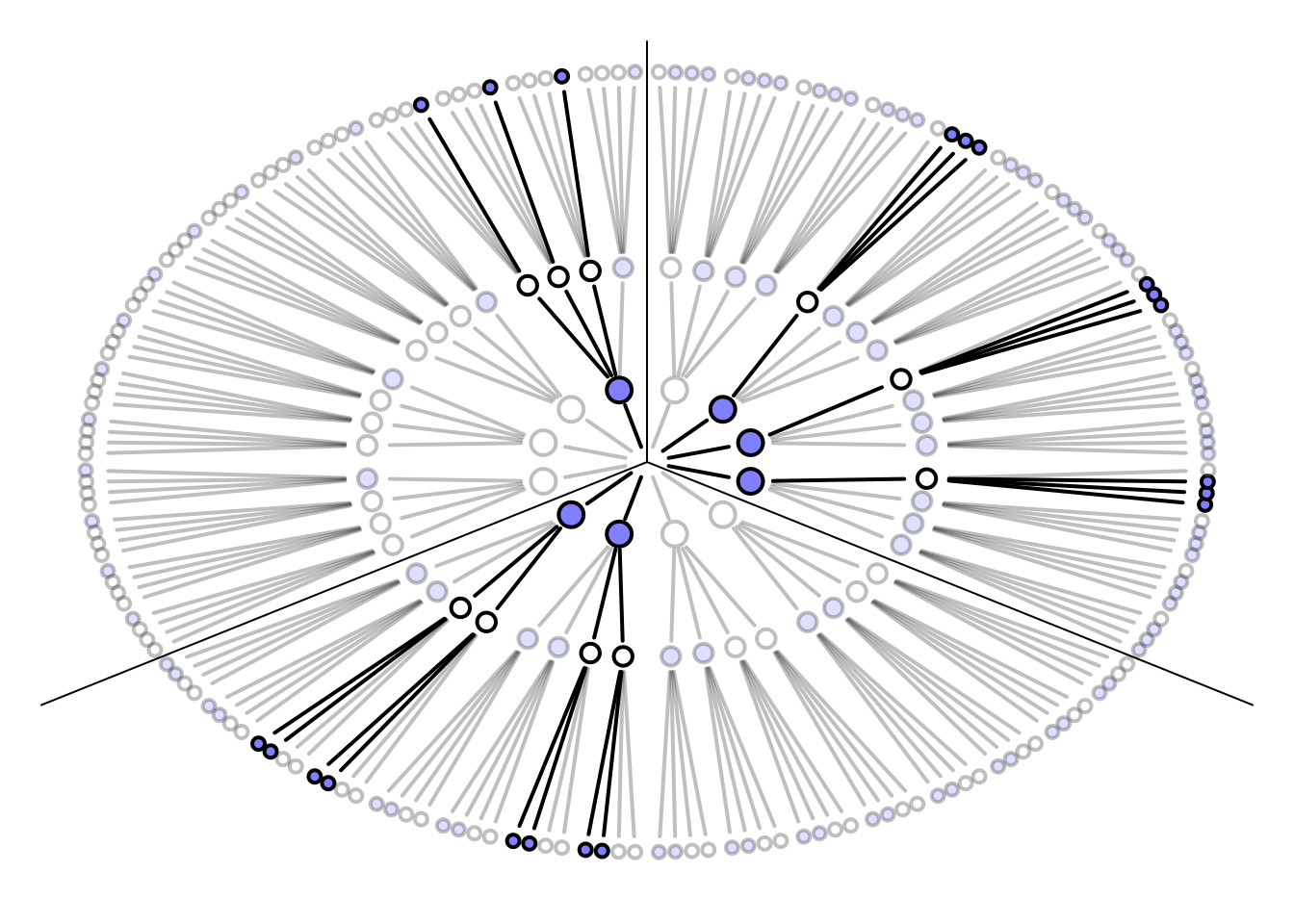
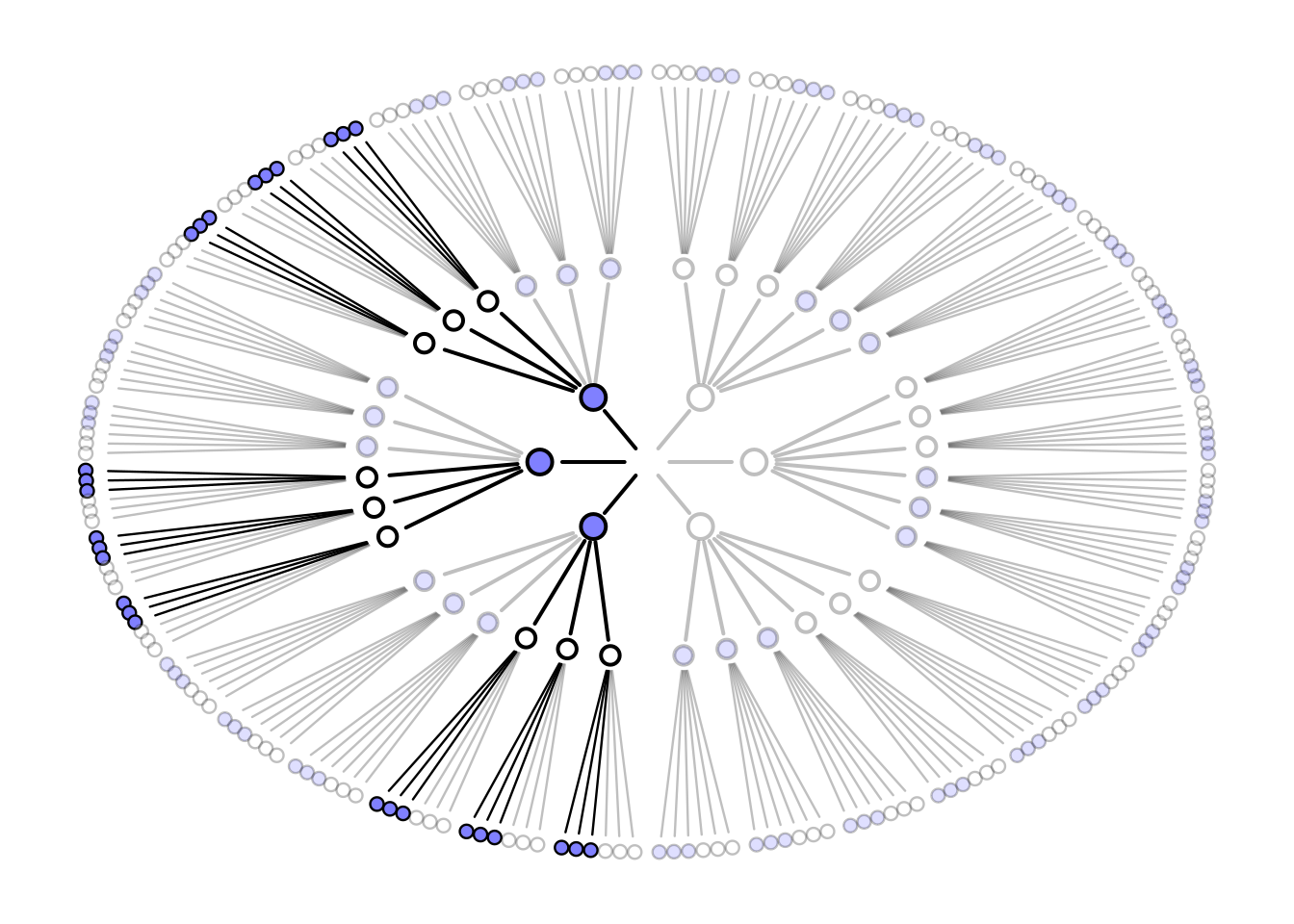
# garden draws paths using recursion
garden <- function( arc , possibilities , data , alpha.fade = 0.25 , hedge=0.1 , hedge1=0 , newplot=TRUE , plot.origin=FALSE , cex=1.5 , lwd=2 , adj.cex , adj.lwd , ring_dist , ... ) {
poss.cols <- ifelse( possibilities==1 , rangi2 , "white" )
if ( missing(adj.cex) ) adj.cex=rep(1,length(data))
if ( missing(adj.lwd) ) adj.lwd=rep(1,length(data))
if ( newplot==TRUE ) {
# empty plot
par(mgp = c(1.5, 0.5, 0), mar = c(1, 1, 1, 1) + 0.1, tck = -0.02)
plot( NULL , xlim=c(-1,1) , ylim=c(-1,1) , bty="n" , xaxt="n" , yaxt="n" , xlab="" , ylab="" )
}
if ( plot.origin==TRUE ) point.polar( 0 , 0 , pch=16 )
N <- length(data)
n_poss <- length(possibilities)
# draw rings
# compute distance out for each ring
# use golden ratio 1.618 for each successive ring
goldrat <- 1.618
if ( missing(ring_dist) ) {
ring_dist <- rep(1,N)
if ( N>1 )
for ( i in 2:N ) ring_dist[i] <- ring_dist[i-1]*goldrat
ring_dist <- ring_dist / sum(ring_dist)
ring_dist <- cumsum(ring_dist)
}
if ( length(alpha.fade)==1 ) alpha.fade <- rep(alpha.fade,N)
draw_wedge <- function(r,hit_prior,arc2,hedge=0.1,hedge1=0,lines_to) {
# is each path clear?
hit <- hit_prior * ifelse( possibilities==data[r] , 1 , 0 )
# transparency for blocked paths
alpha <- ifelse( hit , 1 , alpha.fade[r] )
# draw wedge
hedge_use <- ifelse( r==1 , hedge1 , hedge )
pts <- wedge( ring_dist[r] , arc2[1] , arc2[2] , poss.cols , hedge=hedge_use , alpha=alpha , cex=cex*adj.cex[r] , lwd=lwd*adj.lwd[r] )
if ( N > r ) {
# draw next layer
span <- abs( arc2[1] - arc2[2] ) / n_poss
for ( j in 1:n_poss ) {
# for each possibility, draw the next wedge
# recursion handles deeper wedges
new_arc <- c( arc2[1]+span*(j-1) , arc2[1]+span*j )
pts2 <- draw_wedge(r+1,hit[j],new_arc,hedge,lines_to=pts[j,])
}#j
}# N>r
# draw lines back to parent point
if ( !missing(lines_to) ) {
for ( k in 1:n_poss ) {
alpha_l <- ifelse( hit==1 , 1 , alpha.fade[r] )
line.short( c(lines_to[1],pts[k,1]) , c(lines_to[2],pts[k,2]) , lwd=lwd*adj.lwd[r] , short=0.04 , col=col.alpha("black",alpha_l[k]) )
}
} # lines_to
return(pts)
}
pts1 <- draw_wedge(1,1,arc=arc,hedge=hedge,lines_to=c(0,0))
invisible(pts1)
}
##
goldrat <- 1.618
ring_dist <- rep(1,3)
for ( i in 2:3 ) ring_dist[i] <- ring_dist[i-1]*goldrat
ring_dist <- ring_dist / sum(ring_dist)
ring_dist <- cumsum(ring_dist)
dat <- c(1,0,1)
arc <- c( 0 , pi )
garden(
arc = arc,
possibilities = c(0,0,0,1),
data = dat,
hedge = 0.05,
ring_dist=ring_dist,
alpha.fade=0.35
)